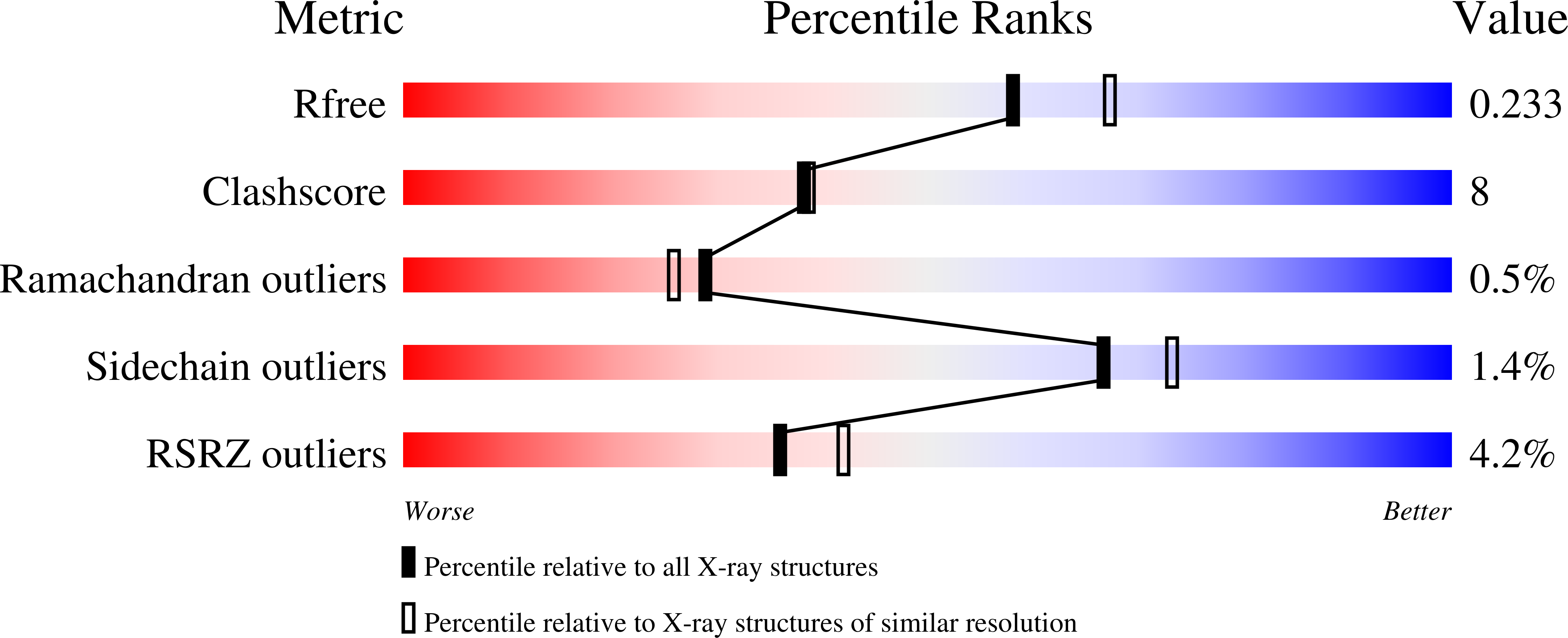
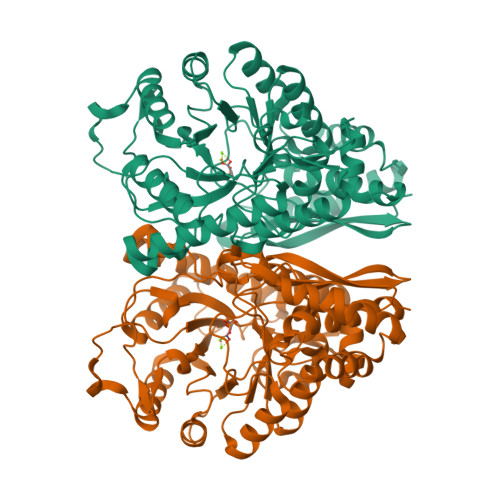
RCSB PDB - 2C7O: HhaI DNA methyltransferase complex with 13mer oligonucleotide containing 2-aminopurine adjacent to the target base (PCGC:GMGC) and SAH

RCSB PDB - 1FUF: CRYSTAL STRUCTURE OF A 14BP RNA OLIGONUCLEOTIDE CONTAINING DOUBLE UU BULGES: A NOVEL INTRAMOLECULAR U*(AU) BASE TRIPLE

RCSB PDB - 1C8C: CRYSTAL STRUCTURES OF THE CHROMOSOMAL PROTEINS SSO7D/SAC7D BOUND TO DNA CONTAINING T-G MISMATCHED BASE PAIRS

RCSB PDB - 1L3U: Crystal Structure of Bacillus DNA Polymerase I Fragment product complex with 11 base pairs of duplex DNA following addition of a dTTP and a dATP residue.
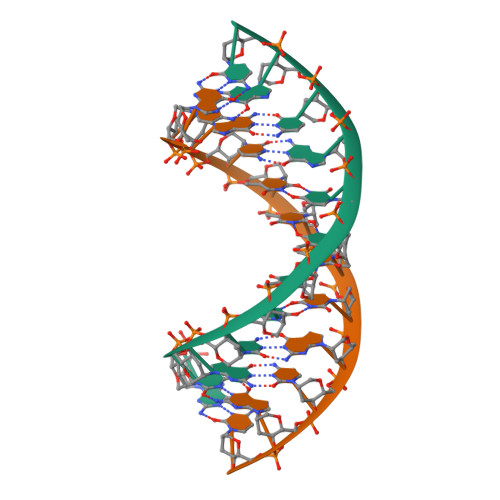
RCSB PDB - 1EC4: SOLUTION STRUCTURE OF A HEXITOL NUCLEIC ACID DUPLEX WITH FOUR CONSECUTIVE T:T BASE PAIRS
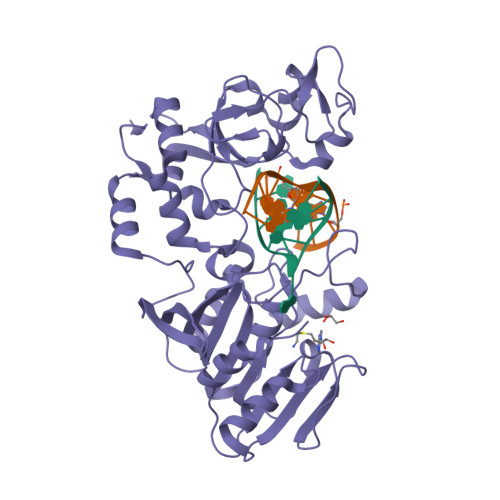
RCSB PDB - 2IH2: Crystal structure of the adenine-specific DNA methyltransferase M.TaqI complexed with the cofactor analog AETA and a 10 bp DNA containing 5-methylpyrimidin-2(1H)-one at the target base partner position

RCSB PDB - 1DA9: ANTHRACYCLINE-DNA INTERACTIONS AT UNFAVOURABLE BASE BASE-PAIR TRIPLET-BINDING SITES: STRUCTURES OF D(CGGCCG)/DAUNOMYCIN AND D(TGGCCA)/ADRIAMYCIN COMPL
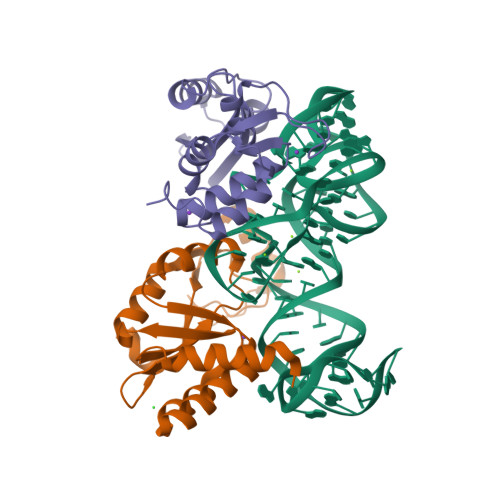
RCSB PDB - 5DAR: CRYSTAL STRUCTURE OF THE BASE OF THE RIBOSOMAL P STALK FROM METHANOCOCCUS JANNASCHII
RCSB PDB - 1BE5: STRUCTURAL STUDIES OF A STABLE PARALLEL-STRANDED DNA DUPLEX INCORPORATING ISOGUANINE:CYTOSINE AND ISOCYTOSINE:GUANINE BASE PAIRS BY NMR, MINIMIZED AVERAGE STRUCTURE

RCSB PDB - 1YFL: T4Dam in Complex with Sinefungin and 16-mer Oligonucleotide Showing Semi-specific and Specific Contact and Flipped Base